Website update to reflect project’s achievements
Blog content
As we enter the last phase of the TRANSGENE project, we have updated our website to reflect the shifts in the way we have been thinking about and studying genomics research. At the outset of the project, we organised our work in terms of the species that individual members of the project were concentrating on – human, yeast and pig – and the work on bibliometric techniques and big data.
These strands, as we called them, represented the way that the project operated in its earlier stages, with individual members of the project team focusing on the trajectory of genomics research of their species, be it human (Miguel García-Sancho), pig (James Lowe) or yeast (Giuditta Parolini and Erika Szymanski). Alongside this, Mark Wong and later Rhodri Leng applied their specialist technical skills to acquiring and cleaning data on DNA sequence submissions and associated publications.
Even then, the strands were never entirely separate. Regular meetings of the project team enabled us to exchange ideas and insights derived from our empirical research, and to discuss the ongoing creation of the dataset that all strands would end up engaging with and using. This process developed further with the advent of collaborative team meetings from autumn 2017 onwards, in the second year of the project.
Ahead of these meetings, the leader of each species strand worked with the datasets and initial network visualisations produced by Mark and Rhodri, and discussed their thoughts, findings and the additional research that was inspired by them with the core project team (James, Miguel and Erika, as well as Mark). This formed the basis of presentations to the wider collaborative team (Ann Bruce, Niki Vermeulen and Gil Viry), who then provided feedback on the burgeoning mixed methods work. This influenced the ongoing shaping and analysis of the datasets and the production of network visualisations.
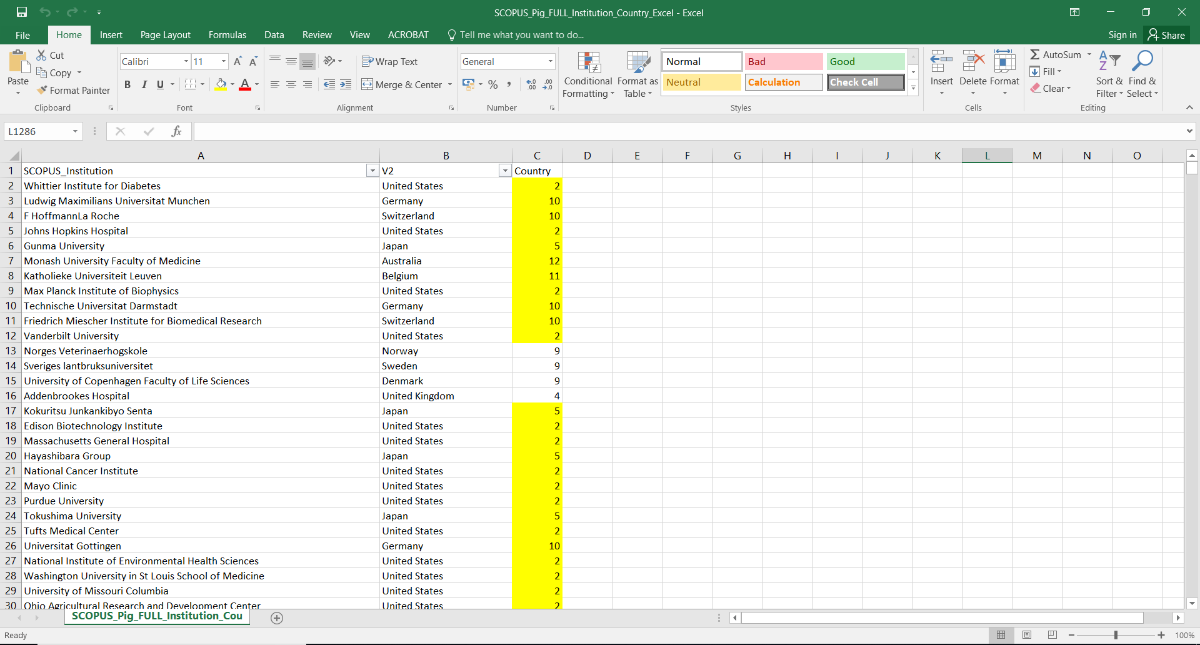
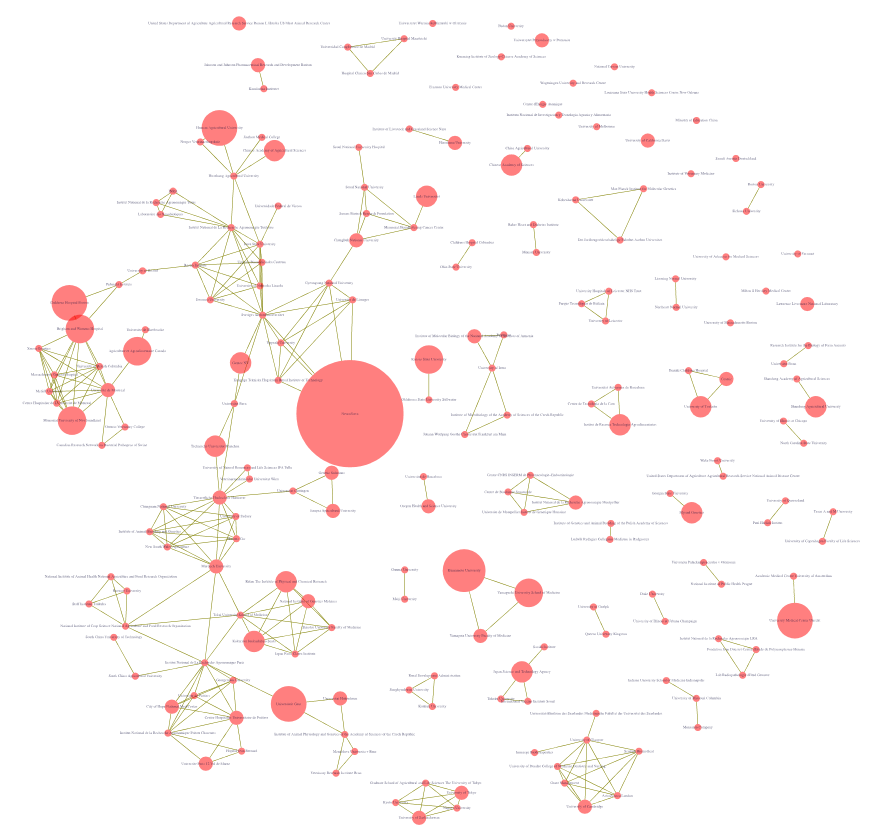
The images above illustrate early datasets (top, data on 2007) and visualisations (institutional co-authorships in 2004) produced in October 2017, concerning institutional data for pig DNA submissions and associated publications.
On this basis, the collaborative team began to organise around the drafting of journal manuscripts using the mixed methods approach that we have developed throughout the project. Meetings would now discuss pre-circulated draft papers, and inspire further quantitative and network-based analysis and qualitative research. Although the analyses of human, pig and yeast genomics have proceeded in distinct ways in these papers, with most members of the collaborative team involved, this process has helped us to draw out connections and themes across the species, and by extension, across the strands.
This, and the insights we have developed within the study of the history of genomics of individual species, has encouraged us to consider the limitations of an approach based on the species categories of DNA sequences submitted to databases such as the European Nucleotide Archive. By doing this, are we merely recapitulating rather than historicising the norms of genomics research itself? Are we missing important facets of genomics research such as the prevalent use of inter-species comparisons? Are we failing to represent modes of research that aim to sample and sequence DNA based on criteria other than it belonging to a given organism of a given species?
Reflecting the shift in the way that we work and our own examination of the shortcomings of a species-bounded approach to studying genomics research, we now articulate our work in terms of broader objectives that we all work towards, rather than as separate strands. These objectives have often been implicitly present in the work we have done. Years of research has now allowed us to flesh them out and state them explicitly, to guide our work until the end of the project.
The objectives are:
- Producing an extensive chronological and analytical history of genomics across three species (yeast, human and pig)
- Combining qualitative and quantitative methods in the historical study of genomics.
Both of these objectives contain an aspiration to be synthetic, and to try to say something about the nature of genomics research and how we can study it, beyond the particularities of the histories of genomics in particular species. As such, they are also intended to help us to shape our research agenda beyond the life of this project, by opening up new vistas in the study of genomics.
