Exploring NHS England’s Genomic Medicine Service through PanelApp
Blog content
By Jarmo de Vries
The author of this blogpost is a PhD student co-supervised by the Principal Investigator and Senior Research Fellow of the TRANSGENE team. The preliminary findings of his doctoral research on the Genomic Medicine Service of NHS England square neatly with the historical trajectory that the TRANSGENE project is uncovering for human genomics research. As much as the Human Genome Project adopted an organisational model that created a rigid distinction between the production and later use of DNA sequence data, the Genomic Medicine Service seems to be pursuing a similar top-down structure in the use of that data for the diagnosis of genetic conditions in the NHS.
In my PhD project, I have been examining the creation of NHS England’s Genomic Medicine Service (GMS). Throughout 2020, I increasingly appreciated the importance of a tool called PanelApp in the handling, interpretation and production of genomic data concerning different genetic conditions. Originally set up by Genomics England for the 100,000 Genomes Project, it has become a key tool in the GMS and through it, links can be found and described between Genomics England, the 100,000 Genomes Project, and the GMS. While talking to a curator at the 2020 Open Meeting of the UK Pharmacogenetics & Stratified Medicine Network (UKPGx), I realised that PanelApp plays a key role in the standardisation of the genomic testing strategy in the GMS.
PanelApp consists of several virtual gene panels for different clinical indications, which can be used for the interpretation of genomic data. The panels have been created and are maintained and updated by the PanelApp curators. They have graded genes green, amber, or red indicating the evidence for the association between the gene and the clinical indication (See Figure 1 for an example panel). The evidence for these grades came from among others existing gene-phenotype databases, expert reviews, and information from diagnostic laboratories.
Specific panels have been developed for interpretation and testing in the GMS. By using these virtual panels, it is possible to look at only those genes assumed relevant to the particular disorder of the patient, thereby decreasing the amount of data that needs to be analysed in the case of whole genome or exome sequencing (WGS/WES). In WGS, the entire DNA is sequenced, while in WES only exons, the protein-encoding parts of genes, are sequenced. In this way, PanelApp is intended to standardise and ease the interpretation of genomic data.
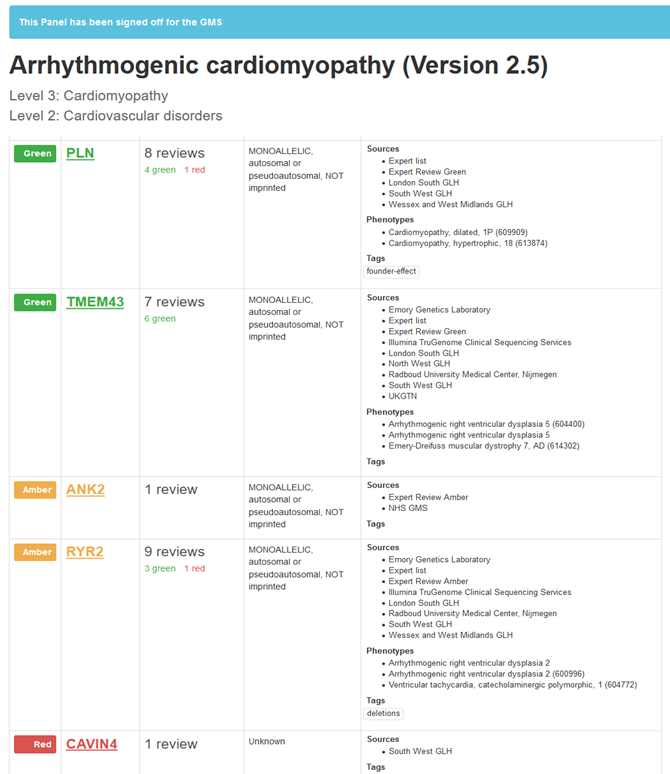
Screenshot from PanelApp: https://panelapp.genomicsengland.co.uk/panels/134/ May 2020
My initial PhD research has focused on how the PanelApp panels have been constructed. I have used the concept of data journeys to map and reconstruct this process, drawing on literature such as Leonelli (2016) and Stevens (2013). Through tracking how genomic data has been moved, used, and reused, I can identify the different actors and trajectories involved in the making of these panels. I can also investigate what PanelApp tells us about the organisation of the GMS. Overall, my mapping was based on my attendance to the UKPGx conference, informal talks with the conference participants, Genomics England documentation on PanelApp, browsing PanelApp itself, and documents from and interviews with people involved in the GMS, such as curators, administrators, and clinical geneticists.
In this blog post, I use PanelApp to show how the implementation of the GMS is creating a new knowledge-control regime of how the genetics services and genomic data are managed. Hilgartner (2017, p. 9) defines knowledge-control regimes as “a sociotechnical arrangement that constitutes categories of agents, spaces, objects, and relationships among them in a manner that allocates entitlements and burdens pertaining to knowledge.” To appreciate its role in the GMS, it is necessary to understand that PanelApp is the result of work done during the 100,000 Genomes Project (100K GP). This project’s goal was to lay the groundwork of the GMS and to enable the transformation of NHS England’s genetics services (as discussed by Samuel and Farsides, 2017). The NHS did not carry out 100K GP; instead, Genomics England, a private company owned by the Department of Health, was set up to lead it.
Following on from 100K GP, the GMS is supposed to fundamentally change NHS genetics services and, with it, the knowledge-control regime associated with them. But what are these changes? First and most important, the GMS centralises and standardises the organisation of genetic testing in the NHS and this becomes visible through the role that PanelApp’s GMS panels have. For each disease, the GMS panels determine what genes will be examined for potential disease-causing variants. These panels have to be used in WGS and WES and in this way they centralise and standardise the genetic testing strategy. As a director of one of the accredited genomic laboratories told me, genetic laboratories had more independence to decide on their testing strategy and organisation of genetic testing before the GMS.
Aside from the apparent centralisation and standardisation, new actors have become involved in the GMS, and the control of genomic data and the testing strategy is not just in the hands of NHS England. Genomics England is the key example of this. PanelApp has been created by them and they will provide the WGS service in the GMS through a partnership with Illumina, a company that manufactures DNA sequencing machines. Furthermore, Genomics England will store data of consenting patients in the GMS in the National Genomic Research Library and will control the access of researchers (with both commercial and non-commercial affiliations) to it.
The PanelApp curators are another good example of new actors in genetics services in the UK. They have primarily created the panels for PanelApp, are responsible for updating and maintaining PanelApp in general, and they are members of the specialist groups that finalise and produce the GMS panels. While I have not yet been able to reconstruct exactly how these specialist groups work, it is likely that the curators have played and will play an important role in producing and revising the GMS panels. This is strengthened by the fact that the GMS panels are partially based on the panels created for 100K GP. Furthermore, PanelApp curators are still updating non-GMS panels and because the process for revising GMS panels is unknown yet, it is likely that updates being made by curators could play an important role in revising the GMS panels. Overall, a picture appears in which genomic laboratories in the GMS cannot independently decide on their testing strategy and, instead, the PanelApp curators arise as important actors in assessing and controlling genomic data, and through the construction of GMS panels as important actors in determining the testing strategy.
This post is a preliminary sketch of some of the developments I perceive to be happening in the GMS. I will continue to research the construction of the GMS panels, their use, and more generally the role of Genomics England in the GMS. What I hoped to have shown, however, is that the introduction of genomic technologies such as WGS and WES and the consequent production and use of huge amounts of genomic data appears to go together with big changes in NHS England’s genetics services. The GMS appears to be provoking a reorganisation of them in which the use, production, and interpretation of genomic data are controlled in a more top-down and centralised fashion.
References
Hilgartner, S., 2017. Reordering Life: Knowledge and Control in the Genomics Revolution. MIT Press, Cambridge, Mass.; London. https://mitpress.mit.edu/books/reordering-life
Leonelli, S., 2016. Data-Centric Biology: A Philosophical Study. University of Chicago Press, Chicago. https://press.uchicago.edu/ucp/books/book/chicago/D/bo24957334.html
Stevens, H., 2013. Life Out of Sequence: A Data-Driven History of Bioinformatics. University of Chicago Press, Chicago. https://press.uchicago.edu/ucp/books/book/chicago/L/bo16744390.html
Samuel, G.N., Farsides, B., 2017. The UK’s 100,000 Genomes Project: manifesting policymakers’ expectations. New Genetics and Society 36, 336–353. https://www.tandfonline.com/doi/full/10.1080/14636778.2017.1370671
